(+1) 302-319-8696
Hieff Canace™ Veritas Long PCR Master Mix (With Dye) _ 10166ES
Size: 250 μL, 1mL, 5x1mL (different size, price is different, please check with sales rep)
Description
Hieff Canace™ Long PCR Master Mix (With Dye) is a ready-to-use 2× pre-mixed solution containing Hieff Canace™ High-Fidelity DNA Polymerase, dNTPs, and an optimized buffer system, which contains pre-added electrophoresis indicators. The pre-mix contains pre-added electrophoresis indicator, PCR products can be directly electrophoresed, the amplification products are flat ends. 2× Hieff Canace™ Long PCR Master Mix (With Dye) has the advantages of quick and easy, high sensitivity, high specificity, good stability, etc., the reaction system can be added with only the primers and templates. In addition, the product also contains a specific protective agent, so that the premix can still maintain stable activity after repeated freezing and thawing.
Ultra-high fidelity: Low mismatch rate in mutation-prone high GC gene;
Fast speed: The speed is as fast as 5 sec/kb;
Wide applicability: It can be used to amplify genes with 20-80% GC content, and tolerate 20% blood and mouse lysates;
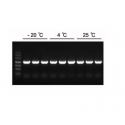
Good stability: Stored at 37°C for 7 days without affecting performance;
Easy use: Just adding primers and templates for amplification, with blue loading dye for direct electrophoresis.
Specifications
| Cat.No. | 10166ES01 | 10166ES03 | 10166ES08 |
| Size | 250 μL | 1 mL | 5×1 mL |
Figures
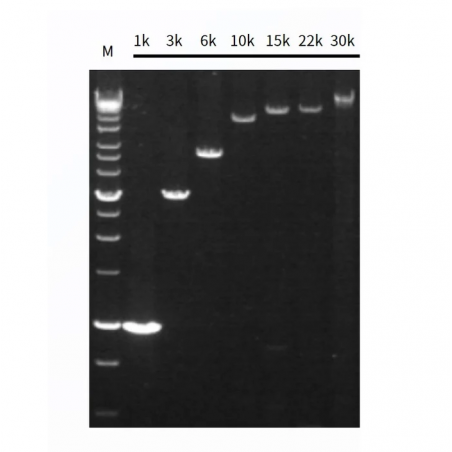
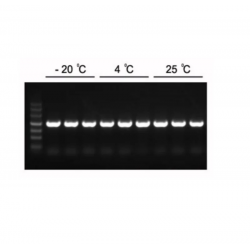
 Figure1. High sensitivity of Hieff Canace™ High-Fidelity Long PCR Master Mix.
Figure1. High sensitivity of Hieff Canace™ High-Fidelity Long PCR Master Mix.

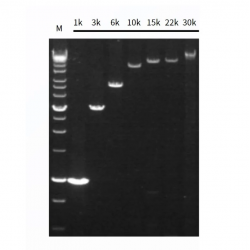
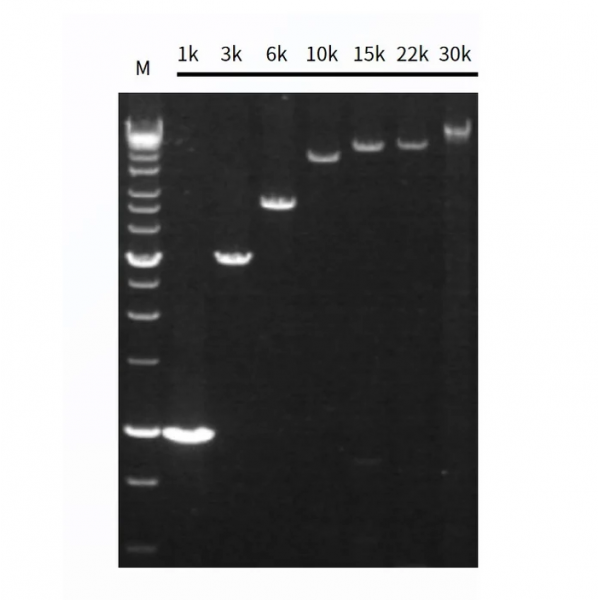
Figure 2. Long PCR from cDNA by Hieff Canace™ High-Fidelity Long PCR Master Mix.
Storage
This product should be stored at -25~-15℃ for 1 years.
Instructions
- Recommended PCR reaction systems.
|
Components |
Volume(μL) |
Final concentration |
|
Hieff Canace™ Long PCR Master Mix (With Dye)* |
25 |
1× |
|
Template** |
x |
- |
|
Forward Primer(10 μmol/L)*** |
2 |
0.4 μmol/L |
|
Reverse Primer(10 μmol/L) |
2 |
0.4 μmol/L |
|
ddH2O |
Up to 50 |
- |
Table 1 PCR reaction system
*In 1× premixes containing 2 mM Mg2+ and 200 μM dNTPs.
**Recommended range 10-200 ng, cDNA sample upload volume range not more than 1/10 of the reaction system, recommended 1-2.5 μL.
***The final primer concentration in the PCR reaction system ranges from 0.2-1 μM, and 0.4 μM is recommended.
- Reaction program.
|
Cycle step |
Temp. |
Time |
Cycles |
|
Initial denaturation |
98℃ |
30 sec |
1 |
|
Denaturation |
98℃ |
10 sec |
30-35 |
|
Annealing* |
60℃ |
5 sec |
|
|
Extension** |
72℃ |
5-10 sec/kb |
|
|
Final extension |
72℃ |
2 min |
1 |
Table 2 PCR reaction program
*Recommended temperature: 60°C, a temperature gradient can be set up to find the optimal temperature for primer annealing. The recommended annealing time is set to 5 sec and can be adjusted from 5-30 sec. Too long annealing time may result in diffuse amplification products on the gel.
**Extension time: Recommended 5 sec/kb, can also be extended to 10 sec/kb as needed.
Notes
- This product is for research use only.
- Please operate with lab coats and disposable gloves,for your safety.
Documents:
Citations & References:
[1] Dong W, Zhu Y, Chang H, et al. An SHR-SCR module specifies legume cortical cell fate to enable nodulation. Nature. 2021;589(7843):586-590. doi:10.1038/s41586-020-3016-z(IF:42.779)
[2] Sun L, Yan Y, Lv H, et al. Rapamycin targets STAT3 and impacts c-Myc to suppress tumor growth. Cell Chem Biol. 2022;29(3):373-385.e6. doi:10.1016/j.chembiol.2021.10.006(IF:8.116)
[3] Ye M, Xiong L, Dong Y, et al. The Potential Role of the Methionine Aminopeptidase Gene PxMetAP1 in a Cosmopolitan Pest for Bacillus thuringiensis Toxin Tolerance. Int J Mol Sci. 2022;23(21):13005. Published 2022 Oct 27. doi:10.3390/ijms232113005(IF:6.208)
[4] He Y, Zhou J, Fu R, et al. The application of DNA-HRP functionalized AuNP probes in colorimetric detection of citrus-associated Alternaria genes. Talanta. 2022;237:122917. doi:10.1016/j.talanta.2021.122917(IF:6.057)
[5] Guo L, Yang W, Huang Q, et al. Selenocysteine-Specific Mass Spectrometry Reveals Tissue-Distinct Selenoproteomes and Candidate Selenoproteins. Cell Chem Biol. 2018;25(11):1380-1388.e4. doi:10.1016/j.chembiol.2018.08.006(IF:5.592)
[6] Dou W, Zhu Q, Zhang M, Jia Z, Guan W. Screening and evaluation of the strong endogenous promoters in Pichia pastoris. Microb Cell Fact. 2021;20(1):156. Published 2021 Aug 9. doi:10.1186/s12934-021-01648-6(IF:5.328)
[7] Xiong Y, Yi Y, Wang Y, Yang N, Rudd CE, Liu H. Ubc9 Interacts with and SUMOylates the TCR Adaptor SLP-76 for NFAT Transcription in T Cells. J Immunol. 2019;203(11):3023-3036. doi:10.4049/jimmunol.1900556(IF:4.718)
[8] Huang L, Xiang M, Ye P, Zhou W, Chen M. Beta-catenin promotes macrophage-mediated acute inflammatory response after myocardial infarction. Immunol Cell Biol. 2018;96(1):100-113. doi:10.1111/imcb.1019(IF:4.557)
[9] Wang X, Du A, Yu G, Deng Z, He X. Guanidine N-methylation by BlsL Is Dependent on Acylation of Beta-amine Arginine in the Biosynthesis of Blasticidin S. Front Microbiol. 2017;8:1565. Published 2017 Aug 22. doi:10.3389/fmicb.2017.01565(IF:4.076)
[10] Yu P, Zhou L, Yang WT, et al. Comparative mitogenome analyses uncover mitogenome features and phylogenetic implications of the subfamily Cobitinae. BMC Genomics. 2021;22(1):50. Published 2021 Jan 14. doi:10.1186/s12864-020-07360-w(IF:3.969)
[11] Liu G, Liu Y, Niu B, et al. Genetic mutation of TRPV2 induces anxiety by decreasing GABA-B R2 expression in hippocampus. Biochem Biophys Res Commun. 2022;620:135-142. doi:10.1016/j.bbrc.2022.06.079(IF:3.575)
[12] Tan KS, Zhang Y, Liu L, et al. Molecular cloning and characterization of an atypical butyrylcholinesterase-like protein in zebrafish. Comp Biochem Physiol B Biochem Mol Biol. 2021;255:110590. doi:10.1016/j.cbpb.2021.110590(IF:2.231)
FAQ
Q: When performing long fragment amplification (over 10kb) with this product, does it still follow the reaction procedure for reactions under 10kb?
A: When dealing with fragments larger than 10 kb, it is advisable to combine 68℃ with 10 sec/kb to enhance the amplification success rate. For fragments smaller than 10 kb, an amplification speed of 5 sec/kb can be used.
Q: Can the 10166ES product be used for the amplification of long cDNA fragments? What suggestions do you have for the experiment?
A: (1) This 10166ES product can accommodate cDNA amplification ranging from 1 kb to 14 kb. If you need to perform long fragment amplification, it is recommended to use the validated 11149ES reverse transcription kit from Yeasen Biotechnology to reverse transcribe the target RNA. The resulting cDNA template can then be used in conjunction with 10166ES for long fragment amplification.
(2) If the RNA sample is small or if you need to obtain a very long target fragment, it is recommended not to perform digestion during reverse transcription. The longer the target fragment, the more RNA needs to be input, and accordingly, more cDNA will be required.
Q: What should we do if there are mutations in the amplified product?
A: 10166ES is a high-fidelity PCR product. High fidelity means a lower error rate and more accurate sequences. However, different experimental methods may result in variations. Product mutations are low-probability events. It is recommended to perform multiple repeated sets of new amplifications to obtain unmutated amplification products.
Q: Can the 10166ES be used to amplify fragments with high GC content?
A: Yes, after testing, the 10166ES can cover fragments with GC content ranging from 20% to 80%. When expanding fragments with a high GC content, it is recommended to adjust the temperature to 68℃, which will significantly increase the success rate.
Q: My sample is a bacterial culture solution. Can I directly use 10166ES to amplify the bacterial culture solution?
A: The 10166ES can tolerate a certain amount of bacterial solution. After testing, it has been found that this product can tolerate bacterial solution at a proportion of 12.5% in the reaction system, and it has good specificity. If you need to directly expand the bacterial solution, please keep the proportion below 12.5%.
Q: Can Hieff Canace®Veritas Long PCR Master Mix (with Dye) be used for amplifying cDNA templates containing uracil?
A: The 10166ES product can tolerate a certain amount of cDNA stock solution, which contains residual RNA and degraded RNA. Therefore, cDNA containing uracil can be used as the template. However, there is a certain range. After product performance testing, 10166ES can meet the requirement of a cDNA input volume accounting for 15% of the reaction system.
Q: How much sample can the Hieff Canace®Veritas Long PCR Master Mix (with Dye) detect?
A: (1) For plant genomes, after product performance testing, it can detect as low as 100pg of plant genome.
(2) For animal genomes, after product performance testing, the minimum detectable amount is 100pg of animal genome.
(3) For plasmids, since they are relatively pure, after product performance testing, the minimum detectable amount is 1pg of plasmid.
Q: Why did I have no problem using 10166ES several times before, but this time when expanding the long fragment, there was no band?
A: Possible causes and solutions:
The thawing requirements for 10166ES stipulate that it should be thawed slowly on ice, and it cannot be thawed at room temperature or through heating.
(2) When performing the amplification process, it is recommended to set the temperature at 68℃ and the amplification speed at 10 seconds per kilobase.
(3) The longer the target fragment to be amplified is, the more template should be used.
(4) In the PCR system, the concentration of primers is too low or the number of cycles is insufficient: appropriately increase the amount of primers, or increase the number of amplification cycles;