(+1) 302-319-8696
Hieff Unicon™ Universal Blue qPCR Master Mix (Dye Based)
Size: 1 mL, 5x1 mL, 50x1 mL, 100x1 mL, (different size price please check with sales rep)
is a pre-solution for 2×real-time quantitative PCR amplification characterized by high sensitivity and specificity, is blue in color, and has the effect of sample addition tracing. The core component Hieff Unicon™ Taq DNA polymerase uses antibody hot start to effectively inhibit non-specific amplification due to primer annealing during sample preparation. At the same time, the formula adds the promoting factors to improve the amplification efficiency of PCR reaction and equalize the amplification of genes with different GC contents (30 ~ 70%), so that quantitative PCR can obtain a good linear relationship in a wide quantitative region. This product contains special ROX Passive Reference Dye, which is applicable to most qPCR instruments. It is not necessary to adjust the concentration of ROX on different instruments. It is only necessary to add primers and templates to prepare the reaction system for amplification.
Features
- Built-in visual indicator: It contains an inert blue dye that does not interfere with real-time PCR and provides higher visibility in tubes or Wells.
- High specificity: It contains Taq DNA polymerase and is tightly controlled by an antibody-mediated heat-priming mechanism, thereby avoiding nonspecific amplification.
- Wide instrument compatibility: It is compatible with most qPCR instruments and eliminates the need to adjust the concentration of ROX on different instruments
Applications
- Gene Expression
Specifications
| Concentration | 2× |
| Detection method | SYBR Green dye |
| PCR method | qPCR |
| Polymerase | Taq DNA polymerase |
| Type of sample | DNA |
| Application equipment | Most qPCR instruments |
| Product Type | Premix for real-time fluorescence quantitative PCR |
| Apply to (application) | Gene Expression |
Components
| Components No. | Name | 11184ES03 | 11184ES08 | 11184ES50 | 11184ES60 |
| 11184 |
Hieff Unicon™ Universal Blue qPCR Master Mix (Dye Based) |
1 mL | 5×1 mL | 50×1 mL | 100×1 mL |
Shipping and Storage
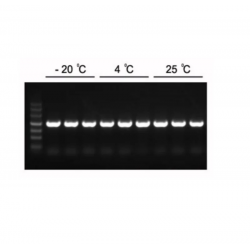
The product is shipped with ice packs and can be stored at -15℃ ~ -25℃ for 18 months. The product contains fluorescent dyes, so it is necessary to avoid strong light irradiation when storing or preparing the reaction system.
Figures
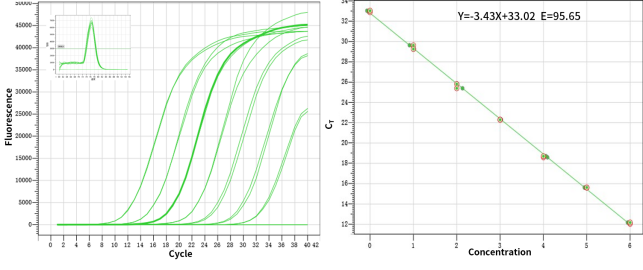
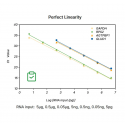
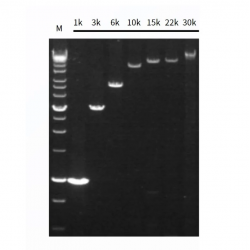
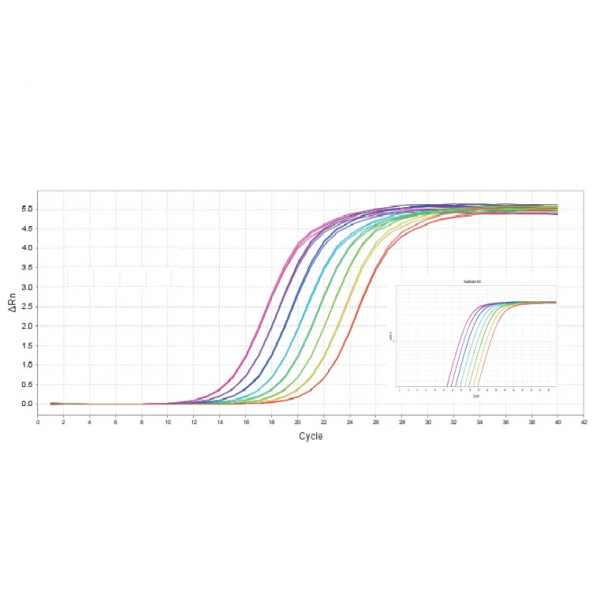
Figure 1. High sensitivity: the ability to detect a single copy.
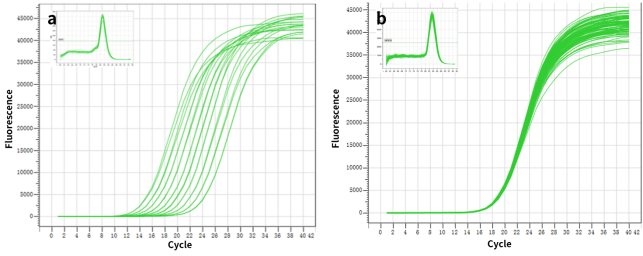
Figure2. High resolution & Excellent reproducibility of duplicate wells.
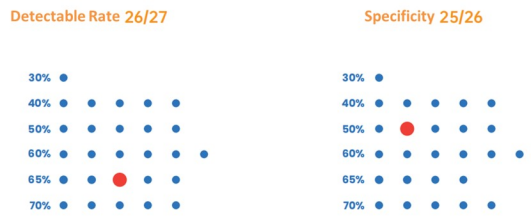
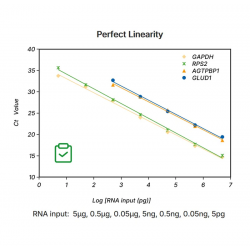
Figure3. Excellent detection rate and specificity: broadly suited for the amplification of 30-70% GC.

Cited from "VCP enhances autophagy-related osteosarcoma progression by recruiting USP2 to inhibit ubiquitination and degradation of FASN, Cell Death & Disease, volume 15, Article number: 788 (2024) "

Cited from "A Defense Pathway Linking Plasma Membrane and Chloroplasts and Co-opted by Pathogens. Cell . 2020 Sep 3;182(5):1109-1124.e25. doi: 10.1016/j.cell.2020.07.020. "
[1] Xia B, Shen X, He Y, et al. SARS-CoV-2 envelope protein causes acute respiratory distress syndrome (ARDS)-like pathological damages and constitutes an antiviral target. Cell Res. 2021;31(8):847-860. doi:10.1038/s41422-021-00519-4(IF:25.617)
[2] Wang S, Li Y, Zhong L, et al. Efficient gene editing through an intronic selection marker in cells. Cell Mol Life Sci. 2022;79(2):111. Published 2022 Jan 31. doi:10.1007/s00018-022-04152-1(IF:9.261)
[3] An LL, Zhao X, Gong XY, et al. Promoter Binding and Nuclear Retention Features of Zebrafish IRF Family Members in IFN Response. Front Immunol. 2022;13:861262. Published 2022 Apr 6. doi:10.3389/fimmu.2022.861262(IF:7.561)
[4] Zhao Y, Wang HP, Yu C, et al. Integration of physiological and metabolomic profiles to elucidate the regulatory mechanisms underlying the stimulatory effect of melatonin on astaxanthin and lipids coproduction in Haematococcus pluvialis under inductive stress conditions. Bioresour Technol. 2021;319:124150. doi:10.1016/j.biortech.2020.124150(IF:7.539)
[5] Shu C, Wang L, Zou C, et al. Function of Foxl2 and Dmrt1 proteins during gonadal differentiation in the olive flounder Paralichthys olivaceus [published online ahead of print, 2022 Jun 16]. Int J Biol Macromol. 2022;215:141-154. doi:10.1016/j.ijbiomac.2022.06.098(IF:6.953)
[6] Wang S, Huang J, Liu F, et al. Isosteviol Sodium Exerts Anti-Colitic Effects on BALB/c Mice with Dextran Sodium Sulfate-Induced Colitis Through Metabolic Reprogramming and Immune Response Modulation. J Inflamm Res. 2021;14:7107-7130. Published 2021 Dec 20. doi:10.2147/JIR.S344990(IF:6.922)
[7] Wang J, Hu R, Wang Z, et al. Establishment of Immortalized Yak Ruminal Epithelial Cell Lines by Lentivirus-Mediated SV40T and hTERT Gene Transduction. Oxid Med Cell Longev. 2022;2022:8128028. Published 2022 Mar 25. doi:10.1155/2022/8128028(IF:6.543)
[8] Liu W, Guan Y, Qiao S, et al. Antiaging Effects of Vicatia thibetica de Boiss Root Extract on Caenorhabditis elegans and Doxorubicin-Induced Premature Aging in Adult Mice. Oxid Med Cell Longev. 2021;2021:9942090. Published 2021 Aug 6. doi:10.1155/2021/9942090(IF:6.543)
[9] Qian Z, Liu C, Li H, et al. Osteocalcin Alleviates Lipopolysaccharide-Induced Acute Inflammation via Activation of GPR37 in Macrophages. Biomedicines. 2022;10(5):1006. Published 2022 Apr 27. doi:10.3390/biomedicines10051006(IF:6.081)
[10] Zhao X, Huang Y, Li X, et al. Full integration of nucleic acid extraction and detection into a centrifugal microfluidic chip employing chitosan-modified microspheres [published online ahead of print, 2022 Jun 27]. Talanta. 2022;250:123711. doi:10.1016/j.talanta.2022.123711(IF:6.057)
FAQ
Q: How many steps are recommended for the qPCR experiment?
A: The common method involves two steps. To enhance amplification specificity, one can choose the two-step method or increase the annealing temperature. When the amplification efficiency is low and the ct value is too high, a three-step method can be adopted or the extension time can be prolonged.
Q: What is the validity of the qPCR experiment results? Why is it recommended that the Ct value should be greater than 15?
A: The validity must meet the following three conditions: (1) Standard curve: Amplification efficiency range: 90-110%, corresponding slope: -3 to -3.5. R2 > 0.98. (Amplification efficiency = 10 - 1/slope - 1), when the slope is -3.32, the amplification efficiency is 100%. (2) Amplification curve: S-shaped curve, and Ct value is between 15 and 35, negative control Ct > 35 or no Ct value. (3) Melting curve: Single peak. The Ct value is greater than 15 cycles because the 10 times standard deviation of the fluorescence value in 3-15 cycles is the fluorescence threshold, and a too small Ct value will affect the curve.
Q: What is the sensitivity limit of 11184?
A: Single copy
Q: Are there differences in the Tm values of the melting curves of different copies of the same gene?
A: Even for the same amplified product, there will be slight differences in the Tm values. Generally, a difference of less than 1 degree is acceptable.
Q: Why did the CT value of the diluted template become smaller instead?
A: Generally, the CT value is negatively correlated with the initial concentration of the template. The higher the concentration, the lower the CT value. However, there are also many special cases. For instance, if there are inhibitors in the system or the template is not pure, diluting the template can actually result in a lower CT value.
Q: The CT value of the internal reference is less than 20, while the target genes are all greater than 30. What should we do?
A: This might be due to the low expression level of the target gene. Suggestions: a) Use a different reference gene; b) Change the primers.
Q: Why is the amplification curve messy and discontinuous?
A: The possible reason is that the ROX was added improperly. Verify whether the reference dye ROX was added correctly.
Q: What is the amount of the template X? What is the commonly used amount?
A: (1) The amount of template DNA needs to be determined by the experimenter during the initial experiment. Firstly, dilute the template DNA (generally recommended to dilute by 5-10 times), then sample at different template quantities on a gradient, and select the optimal sample amount with a CT value ranging from 20 to 30.
(2) The commonly used amount is 500-1000 ng of total RNA for reverse transcription, dilute it 10 times and take 1 μL of cDNA for the qPCR experiment.
Q: What is the validity of the qPCR experimental results? Why is it recommended that the Ct value should be greater than 15?
A: a) The validity must meet the following three conditions:
(1) Standard curve: Amplification efficiency range: 90-110%, corresponding slope: -3 to -3.5. R2 > 0.98. (Amplification efficiency = 10 - 1/slope - 1), when the slope is -3.32, the amplification efficiency is 100%.
(2) Amplification curve: S-shaped curve, and Ct value is between 15 and 35, negative control Ct > 35 or no Ct value.
(3) Melting curve: Single peak.
b) The 10 times the standard deviation of the fluorescence values for 3 to 15 cycles is the fluorescence threshold. A too small Ct value will affect the curve.
Q: What is the function of Rox?
A: ROX is a reference dye. Its function is to standardize the non-PCR oscillations in the fluorescence quantitative reaction, correct the sample addition errors or the errors between wells, and provide a stable baseline.
Q: Why does the amplified product show a tailing effect when running on the gel?
A: The reagent contains stabilizing components that do not participate in any reactions themselves, but they will show a diffused pattern on the electrophoresis gel. It is not recommended to perform electrophoresis after the run is complete.
Q: The reagents are rather thick and prone to forming bubbles. How can we prevent this?
A: You can first mix the large sample, then add it separately to the centrifuge tubes, and finally add the template.
When adding samples to the gun head, make sure not to let any air in. Use the second gear for sample aspiration and the first gear for sample injection; slowly push out the liquid and do not blow forcefully; guide the liquid into the hole along the wall.
After the configuration is completed, if there are still bubbles, remove the centrifuged PCR tube. If it is a 96-well plate, gently tap the 96-well plate after the pipetting is done.